Minimise, Maximise
(sitting at SKA)
Struggling to get back into my code.
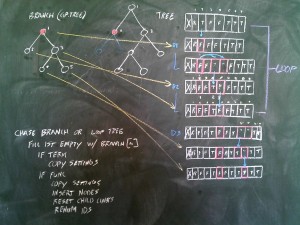
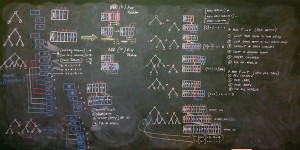
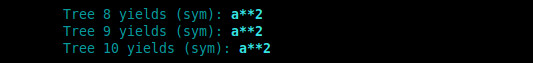
Preparing to test Kepler’s 3rd Law of Planetary Motion. This is a minimisation problem, meaning the best overall fitness will be the smallest number. I will employ the Absolute Value Difference fitness function I recently developed. Simple in theory, but always a few hidden challenges in implementation.
(later)
Realised I need to re-think all my fitness functions, and embed both minimisation and maximisation into the tournament and ‘fitness gym’. Will move to define each fitness kernel as ‘min’ or ‘max’ at the opening.